Phylogeny, genetic diversity and divergence dating of Monodactylus argenteus (Linnaeus, 1758) (Actinopterygii: Monodactylidae) from marine waters of Odisha Coast, Bay of Bengal, India
Abstract
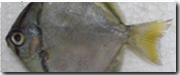
Among the many different types of aquatic life found in marine ecosystems, fish are the most diverse and commercially important organisms. To support their conservation and management, accurate species identification, genetic, and phylogenetic association studies are crucial. Monodactylus argenteus, the silvery Moony fishes were collected from Gopalpur-on-sea, Odisha Coast of the Bay of Bengal, India and identified as using traditional morpho-taxonomy methods followed by DNA barcoding using cytochrome c oxidase subunit I (COI) gene. Following identification, the phylogeny, genetic diversity, and divergence time of M. argenteus were investigated. The current study looked at the number of variable sites, parsimony informative sites, nucleotide diversity, and haplotype diversity. The 16 sequenced individuals of M. argenteus produced a total of 13 haplotypes, with 11 unique haplotypes and two shared haplotypes. There were 67 polymorphic sites, including 56 parsimony informative sites and 11 singleton variable sites with 72 mutations. Phylogenetic tree was drawn and all the sequences clustered in agreement with their species level taxonomic classification were observed. The divergence time of the M. argenteus species was estimated to be in the late oligocene sub-epoch, about 25.98 mya, using the RelTime maximum likelihood method. The findings of this study serve as noteworthy confirmation of the utility of DNA barcode sequences for tracking diversity of species and also contribute information on the phylogeny, genetic diversity, and divergence dating of M. argenteus.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215(3): 403–410.
Anjum S, Ilahi I, Zaman Q, Shah SN, Abbas M, ... Khan H (2025) DNA barcoding, phylogenetics, and morphometric analysis of various freshwater fishes. Journal of Freshwater Ecology 40(1): 2465415.
Ardura A, Planes S, Garcia-Vazquez E (2013) Applications of DNA barcoding to fish landings: authentication and diversity assessment. Zookeys 365: 49–65.
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16(1): 37–48.
Barik TK, Swain SN, Sahu B, Tripathy B, Acharya UR (2018) Morphological and genetic analyses of the first record of longrakered trevally, Ulua mentalis (Perciformes: Carangidae) and of the pinjalo snapper, Pinjalo pinjalo (Perciformes: Lutjanidae) in the Odisha coast, Bay of Bengal. Mitochondrial DNA Part A 29(4): 552–560.
Barik TK, Swain SN, Sahu B, Tripathy B, Acharya UR (2020) Morphological and molecular evidence supports the first occurrence of two fishes, Siganus sutor (Valenciennes, 1835) and Seriolina nigrofasciata (Rüppell, 1829) (Actinopterygii: Perciformes), from marine waters of Odisha coast, Bay of Bengal, India. Acta Oceanologica Sinica 39(6): 26–35.
Barik TK, Swain SN, Sahu B, Tripathy B, Acharya UR (2021) Molecular evidence for Myripristis jacobus and Scarus taeniopterus new to Bay of Bengal: Sporadic appearance or preliminary colonization? Marine Ecology 42: e12632.
Bernatchez L (1991) A role for molecular systematic in defining evolutionary significant units in fishes (pp. 114–132). In: Nielsen JL (Ed) Evolution and the aquatic ecosystem: defining unique units in population conservation. American Fisheries Society, Bethesda, Maryland.
Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, ... Orti G (2017) Phylogenetic classification of bony fishes. BMC Evolutionary Biology 17(162): 1–40.
Bingpeng X, Heshan L, Zhilan Z, Chunguang W, Yanguo W, Jianjun W (2018) DNA barcoding for identification of fish species in the Taiwan Strait. PLoS One 13(6): e0198109.
Erwin DH (1989) Molecular clocks, molecular phylogenies and the origin of phyla. Lethaia 22(3): 251–257.
Fricke R, Eschmeyer WN, Van Der Laan R (2022) Eschmeyer's catalog of fishes: genera, species, references. Electronic version accessed on 15.12.2022.
Fricke R, Eschmeyer WN, Van Der Laan R (2025). Eschmeyer's catalog of fishes: genera, species, references. Electronic version accessed on 24.06.2025.
Froese R, Pauly D (2025). FishBase. World Wide Web electronic publication. www.fishbase.org (Accessed in April 2025).
Fu Y, Li W (1993) Statistical tests of neutrality of mutations. Genetics 133: 693–709.
Gavrilets S (2003) Perspective: models of speciation: what have we learned in 40 years? Evolution 57(10): 2197–2215.
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
Hasegawa M, Kishino H, Yano T (1985) Dating the human-ape split by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 22: 160–174.
Hebert PDN, Cywinska A, Ball SL, de Waard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313–321.
Hillis DM, Huelsenbeck JP, Cunningham CW (1994) Application and accuracy of molecular phylogenies. Science 264(5159): 671–677.
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN (2007) Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes 7(4): 544–548.
Jukes TH, Cantor CR (1969) Evolution of protein molecules (pp. 21–132). In: Monro HN (Ed) Mammalian protein metabolism, Academic Press, Elsevier.
Kaltz O, Shykoff J (1998) Local adaptation in host-parasite systems. Heredity 81(4): 361–370.
Kirkpatrick M, Barton NH (1997) Evolution of a species range. The American Naturalist 150(1): 1–23.
Kumar S, Steche, G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549.
Lange M, Wang H, Zhihong H, Jehle JA (2004) Towards a molecular identification and classification system of lepidopteran-specific baculoviruses. Virology 325(1): 36–47.
Levene H (1953) Genetic equilibrium when more than one ecological niche is available. The American Naturalist 87(836): 331–333.
Manktelow M (2010) History of taxonomy Volume 29. Lecture from Department of Systematic Biology, Uppsala University, Uppsala.
Mohapatra A, Mohanty RK, Mohanty SK, Bhatta KS, Das NR (2007) Fisheries enhancement and biodiversity assessment of fish, prawn and mud crab in Chilika lagoon through hydrological intervention. Wetland Ecology and Management 15(3): 229–251.
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York.
Nei M, Tajima F (1983) Maximum likelihood estimation of the number of nucleotide substitutions from restriction sites data. Genetics 105: 207–217.
Nelson JS (1994) Fishes of the world, third edition. John Wiley & Sons, Inc., New York. 600 pp.
Palumbi SR, Warner RR (2003) Why gobies are like hobbits. Science 299(5603): 51–52.
Patil TS, Tamboli AS, Patil SM, Bhosale AR, Govindwar SP, Muley DV (2016) Relative profile analysis of molecular markers for identification and genetic discrimination of loaches (Pisces, Nemacheilidae). Comptes Rendus Biologies 339(9–10): 364–370.
Pisani D, Benton MJ, Wilkinson M (2007) Congruence of morphological and molecular phylogenies. Acta Biotheoretica 55(3): 269–281.
Rabosky DL, Chang J, Title PO, Cowman PF, Sallan L, ... Alfaro ME (2018) An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559(7714): 392–395.
Rabosky DL, Santini F, Eastman J, Smith SA, Sidlauskas B, ... Alfaro ME (2013) Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Communications 4: 1958.
Rasmussen RS, Morrissey MT, Hebert PDN (2009) DNA barcoding of commercially important Salmon and Trout species (Oncorhynchus and Salmo) from North America. Journal of Agricultural and Food Chemistry 57(18): 8379–8385.
Romano SL, Cairns SD (2000) Molecular phylogenetic hypotheses for the evolution of scleractinian corals. Bulletin of Marine Science 67(3): 1043–1068.
Saarman NP, Louie KD, Hamilton H (2010) Genetic differentation across eastern Pacific oceanographic barriers in the threatened seahorse Hippocampus ingens. Conservation Genetics 11: 1989–2000.
Sambrook J, Russell RW (2001) Molecular cloning: a laboratory manual, 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Stajich JE, Hahn MW (2005) Disentangling the effects of demography and selection in human history. Molecular Biology and Evolution 22(1): 63–73.
Suárez-Díaz E, Anaya-Muñoz VH (2008) History, objectivity, and the construction of molecular phylogenies. Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences 39(4):451–468.
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3): 585–595.
Talwar PK, Kacker RK (1984) Commercial sea fishes of India. Zoological Survey of India, Calcutta. 997 pp.
Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S (2012) Estimating divergence times in large molecular phylogenies. Proceedings of the National Academy of Sciences 109: 19333–19338.
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10 (3): 512–526.
Tamura K, Qiqing T, Kumar S (2018) Theoretical foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Molecular Biology and Evolution 35: 1770–1782.
Ugwumba A, Ugwumba A (2003) Aquaculture options and the future of fish supply in Nigeria. Zoologist, 2(2): 96–122.
Vartak VR, Narasimmalu R, Annam PK, Singh DP, Lakra WS (2015) DNA barcoding detected improper labelling and supersession of crab food served by restaurants in India. Journal of the Science of Food and Agriculture 95(2): 359–366.
Wang SSW, Jinxia J, Xianguang M, Zhongming W, Xiaoyu K (2014) Applicability of mitochondrial COI and 16S rRNA gene sequences in species identification of sole fish (Pleuronectiformes: Soleidae). Tropical Oceanography 33(3): 8.
Ward RD (2000) Genetics in fisheries management. Hydrobiology 420: 191–201.
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences 360(1462): 1847–1857.
Zhang J, Hanner R (2012) Molecular approach to the identification of fish in the South China Sea. PLoS ONE 7(2): e30621.
Copyright (c) 2025 The Author(s)

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.





